Vitamin D blood concentration: phenotype description
Yan Holtz, Zhihong Zhu, Julanne Frater, Perry Bartlett, Jian Yang, John McGrath
This file is the first part of the vitamin-D GWAS analysis. What it does:
- Create a Phenotype file with vitamin-D + other traits of interest like sex, BMI, height, vitaminD nutrition
- Describe Vitamin-D dataset: seasonality, distribution etc.
- Create a Phenotype file with the IPAQ Score of UKB people
# We will need a few library to run this document
library(tidyverse)
library(rmarkdown) # You need this library to run this template.
library(epuRate) # Install with devtools: install_github("holtzy/epuRate", force=TRUE)
library(xtable)
library(knitr)Describe Vitamin-D
#Load the file in R
data <- read.table("0_DATA/pheno.txt.gz", header=T, sep=" ")Dimension
tot <- nrow(data)
blood <- nrow(data[which(!is.na(data$f.4079.0.0)),])I have 502629 people in the UKB.
Among them 468182 people have info concerning their blood pressure (93.146635 %)
Distribution
Distribution of the distolic blood pressure:
data %>%
ggplot( aes(x=f.4079.0.0)) +
geom_density( fill="skyblue", color="skyblue") +
xlab("Diastolic blood pressure")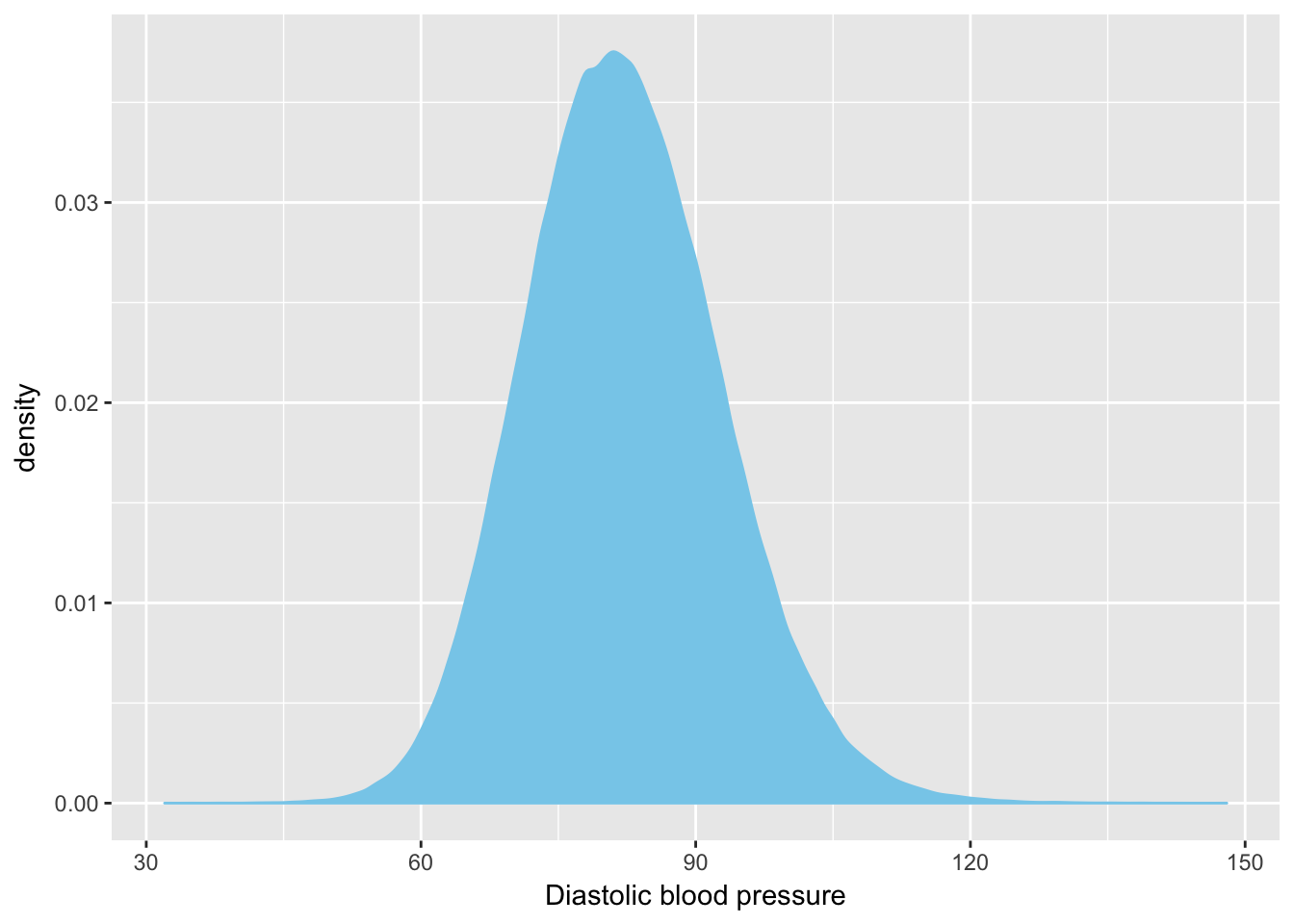
IPAQ score phenotype
The IPAQ score is an estimate of outdoor activities. (International Physical Activity Questionnaire Score). It takes into account:
- Walking: Number of days/week walked 10+ minutes (Field 864.00) and Duration of walk (field 874.00).
- Moderate: Number of days/week of moderate physical activity 10+ minutes (Field 884.00) and Duration of moderate activity (Field 894.00).
- Vigorous: Number of days/week of vigorous physical activity 10+ minutes (Field 904.00) and Duration of vigorous activity (Field 914.00).
The sum of them gives the IPAQ score. Let’s compute it in R.
This work is done on the Inode server, the file I get is transfered locally using ssh.
# good repertory
ssh uqyholtz@inode.qbi.uq.edu.au
cd /ibscratch/wrayvisscher/Yan_Holtz/4_UKB_GWAS/3_BLOOD_GWAS/0_DATA
# Get the good column + repeat the ID + change header to respect plink format
zcat /references/UKBiobank/pheno/download/9280_12505_UKBiobank.tab.gz | awk '{print $1,$1,$8,$22,$280,$283,$286,$289,$292,$295}' > tmp
# Transfert locally
cd /Users/y.holtz/Desktop
scp uqyholtz@inode.qbi.uq.edu.au:/ibscratch/wrayvisscher/Yan_Holtz/0_DATA/tmp .
# Open this file in R, and compute IPAQ
R
data <- read.table("tmp.gz", header=T, sep=" ")
a <- data$`f.864.0.0` * data$`f.874.0.0` * 3.3
b <- data$`f.884.0.0` * data$`f.894.0.0` * 4
c <- data$`f.904.0.0` * data$`f.914.0.0` * 8
data$IPAQ <- a + b + c
colnames(data)[c(1,2)] <- c("FID", "IID")
# Number of missing data?
length(which(is.na(data$IPAQ)))
# Save with new score
write.table(data, file="UKB_IPAQ_score.txt", row.names=FALSE, quote=FALSE)
# Send it on Delta
scp UKB_IPAQ_score.txt y.holtz@delta.imb.uq.edu.au:/shares/compbio/Group-Wray/YanHoltz/DATA/PHENOTYPE/UKBA work by Yan Holtz
Yan.holtz.data@gmail.com