Application of mendelian randomization methods using methylation data
Yan Holtz, Zhihong Zhu, Julanne Frater, Perry Bartlett, Jian Yang, John McGrath
Method
I’m gonna use the methylation data located on Delta at /gpfs/gpfs01/polaris/Q0286/uqywu16/omics/methy
I run SMR on it, like I’ve done for eQTL data.
DNA methylation data description
# load data
data <- read.table("0_DATA/smr_methyl_VitaminDXiaEtAl.smr", header=T)The data set is composed by 85334 SNP - gene methylation associations. These association are distributed along every chromosomes:
table(data$ProbeChr) %>%
as.data.frame() %>%
ggplot( aes(x=Var1, y=Freq)) +
geom_bar(stat="identity", fill="skyblue") +
xlab("chromosome") +
ylab("Number of SNP / gene methylation association") +
theme_ipsum()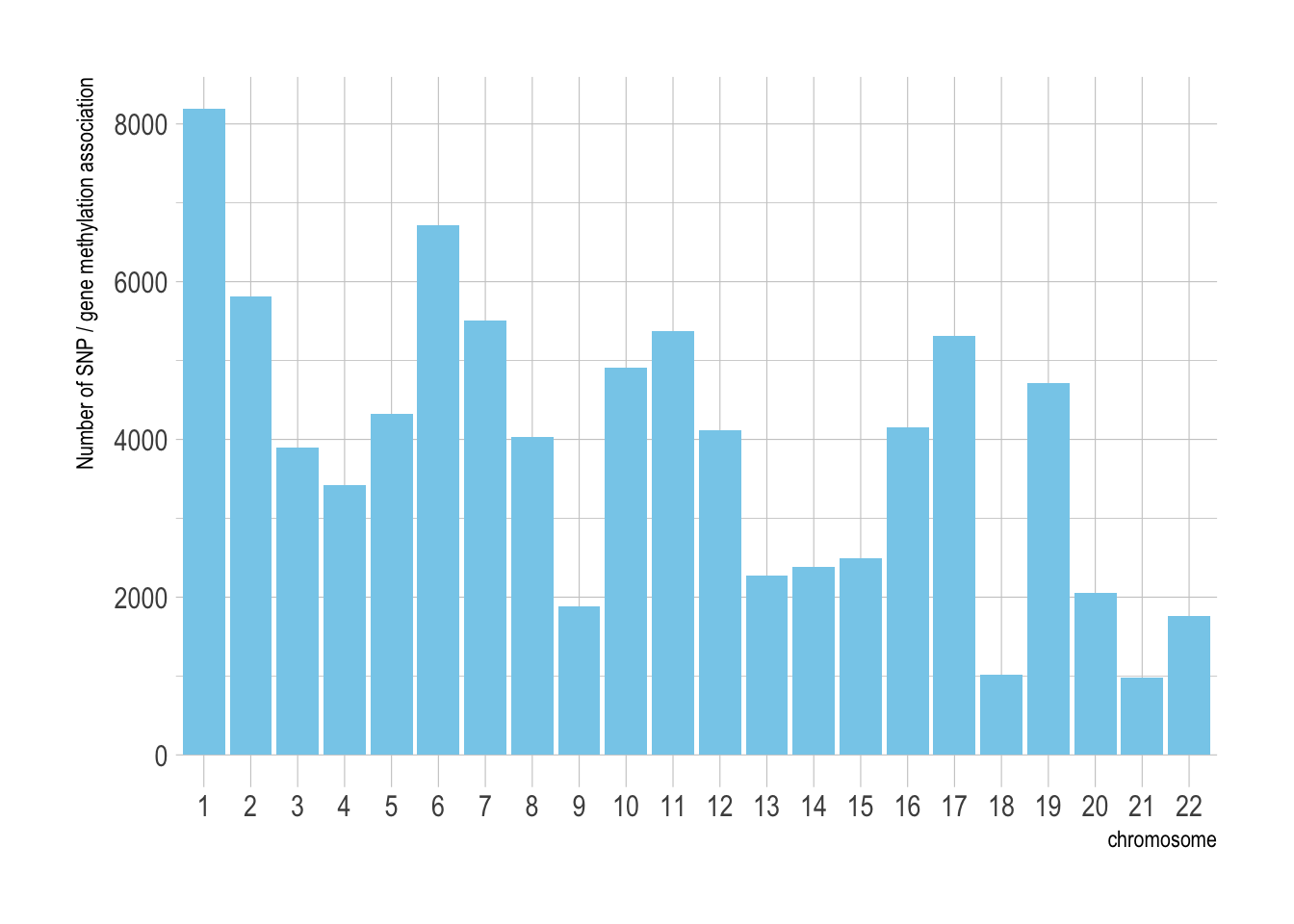
Run SMR
The first step of SMR consists to run the algorythm on every significant mQTL (mQTL = significant relationship between a SNP allele and the methylation of a gene). For each gene involved in an mQTL, we are going to test its putative effect on our trait (vitaminD) thanks to the GWAS summary statistic of this trait.
# Good directory
cd /shares/compbio/Group-Wray/YanHoltz/VITAMIND_XIA_ET_AL/9_METHYLATION
# Run the analysis
tmp_command="smr_Linux --bfile /gpfs/gpfs01/polaris/Q0286/UKBiobank/v2EURu_HM3/ukbEURu_imp_chr{TASK_ID}_v2_HM3_QC --gwas-summary /shares/compbio/Group-Wray/YanHoltz/VITAMIND_XIA_ET_AL/1_GWAS/GWAS_vitaminD_XiaEtAL.ma --beqtl-summary /gpfs/gpfs01/polaris/Q0286/uqywu16/omics/methy/bl_meqtl_cis_std_chr{TASK_ID} --out smr_methyl_VitaminDXiaEtAl_{TASK_ID} --thread-num 1"
qsubshcom "$tmp_command" 1 30G smr_VitaminD 10:00:00 "-array=1-22"
# Concatenate chromosome results
cat smr_methyl_V*smr | head -1 > smr_methyl_VitaminDXiaEtAl.smr
cat smr_methyl_V*smr | grep -v "^probeID" >> smr_methyl_VitaminDXiaEtAl.smr
# Transfer locally
cd /Users/y.holtz/Dropbox/QBI/4_UK_BIOBANK_GWAS_PROJECT/VitaminD-GWAS/0_DATA
scp y.holtz@delta.imb.uq.edu.au:/shares/compbio/Group-Wray/YanHoltz/VITAMIND_XIA_ET_AL/9_METHYLATION/smr_methyl_VitaminDXiaEtAl.smr .Result
Significant SMR results are found in 3 genetic regions. These 3 regions are also detected by the Vitamin D GWAS.
Threshold used for significance: 0.05 / # of Methylation association = 0.05 / 85334 = 5.859329210^{-7}
# Compute signif threshold
thres <- 0.05 / nrow(data)Chromo 4 (GC)
# Show significant association:
data %>%
filter(p_SMR < thres & topSNP_chr==4)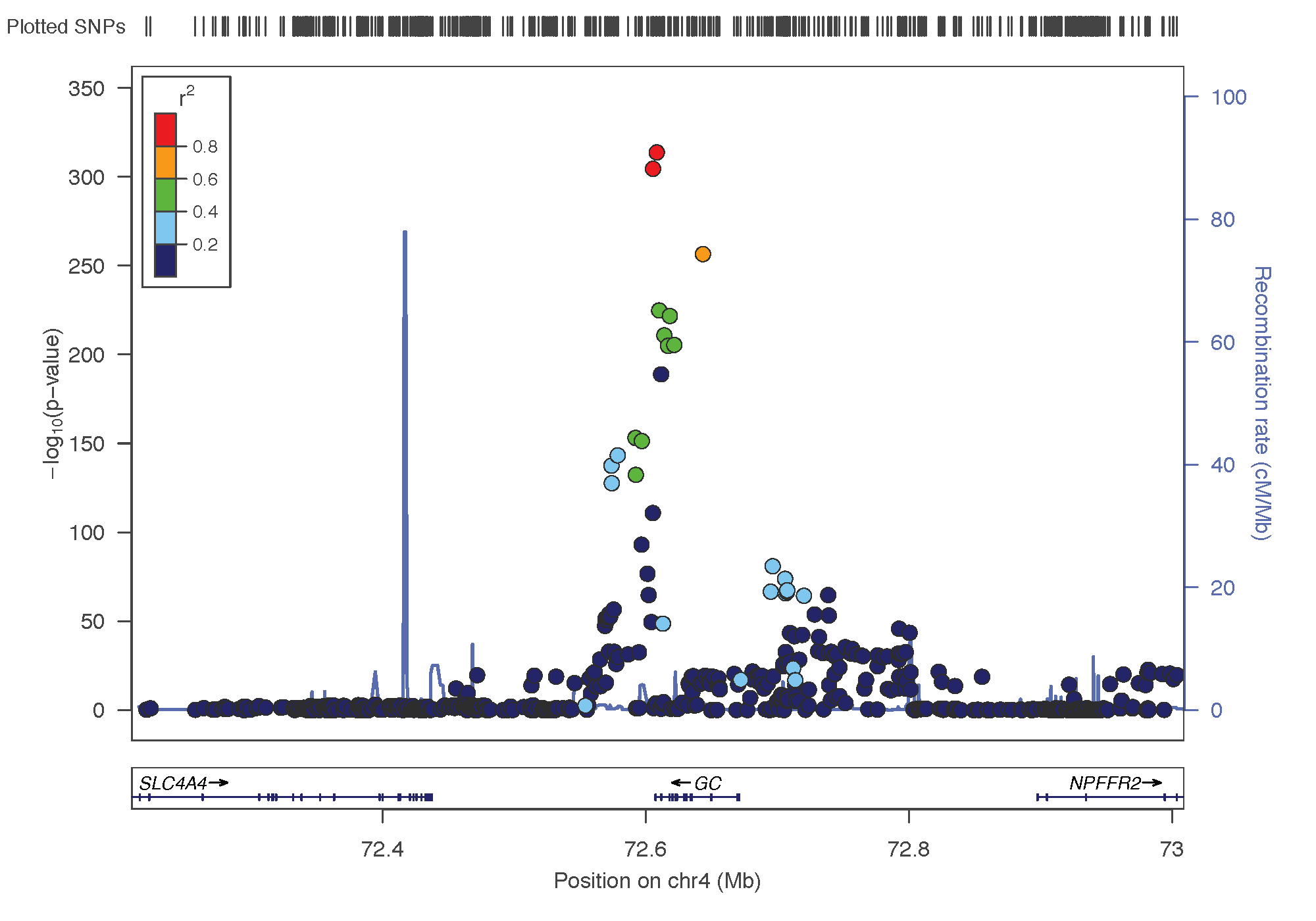
Can I make a diagram of these associations?
tmp <- data %>%
filter(ProbeChr==4) %>%
filter(Probe_bp>71000000 & Probe_bp<74000000) %>%
dplyr::select(Gene, topSNP, p_SMR, Probe_bp, topSNP_bp) %>%
gather(key, position, -1, -2, -3) %>%
mutate(position=position/1000000)
tmp %>% ggplot( aes(y=position, x=key)) +
geom_line(aes(group=paste(Gene,topSNP), color=ifelse(p_SMR<thres,"signif","non signif"))) +
geom_point() +
geom_text(
data=subset(tmp, key=="Probe_bp") %>% dplyr::select(Gene, key, position) %>% group_by(Gene) %>% summarize(key=unique(key), position=mean(position)),
aes( label=Gene) , angle=45, color="grey", hjust=1, nudge_x = -0.1
) +
geom_text(
data=subset(tmp, key=="topSNP_bp") %>% dplyr::select(topSNP, key, position) %>% group_by(topSNP) %>% summarize(key=unique(key), position=mean(position)),
aes( x=key, label=topSNP) , angle=45, color="grey", hjust=0, nudge_x = 0.1
) +
coord_flip() +
theme_ipsum() +
xlab("") +
ylab("Position (Mb)") +
labs(color="SMR assoc with Vitamin-D")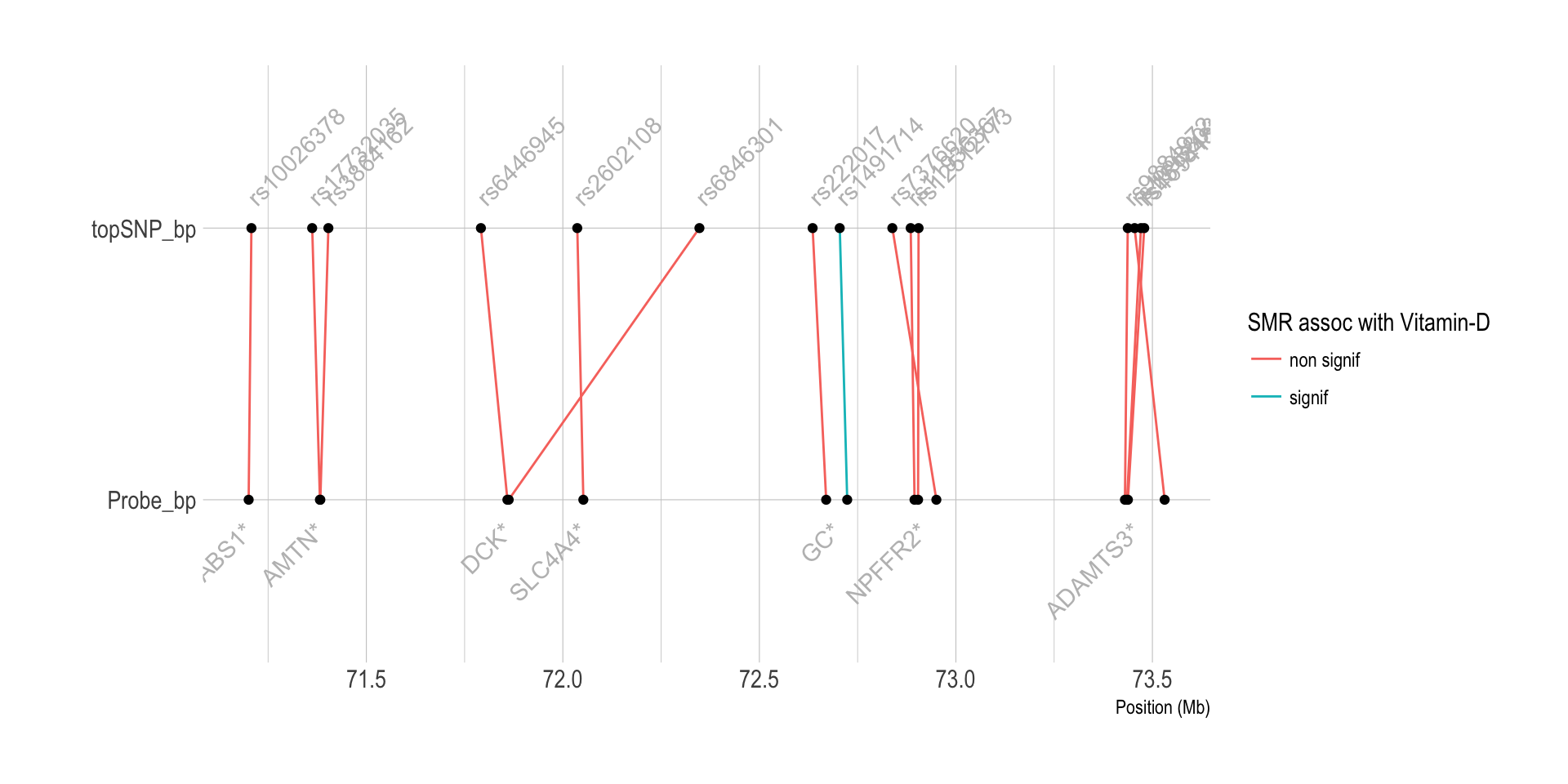
Chromo 11 ~ 14Mb (CYP2R1)
# Show significant association:
data %>%
filter(p_SMR < thres & topSNP_chr==11 & topSNP_bp<40000000) %>%
arrange(Probe_bp)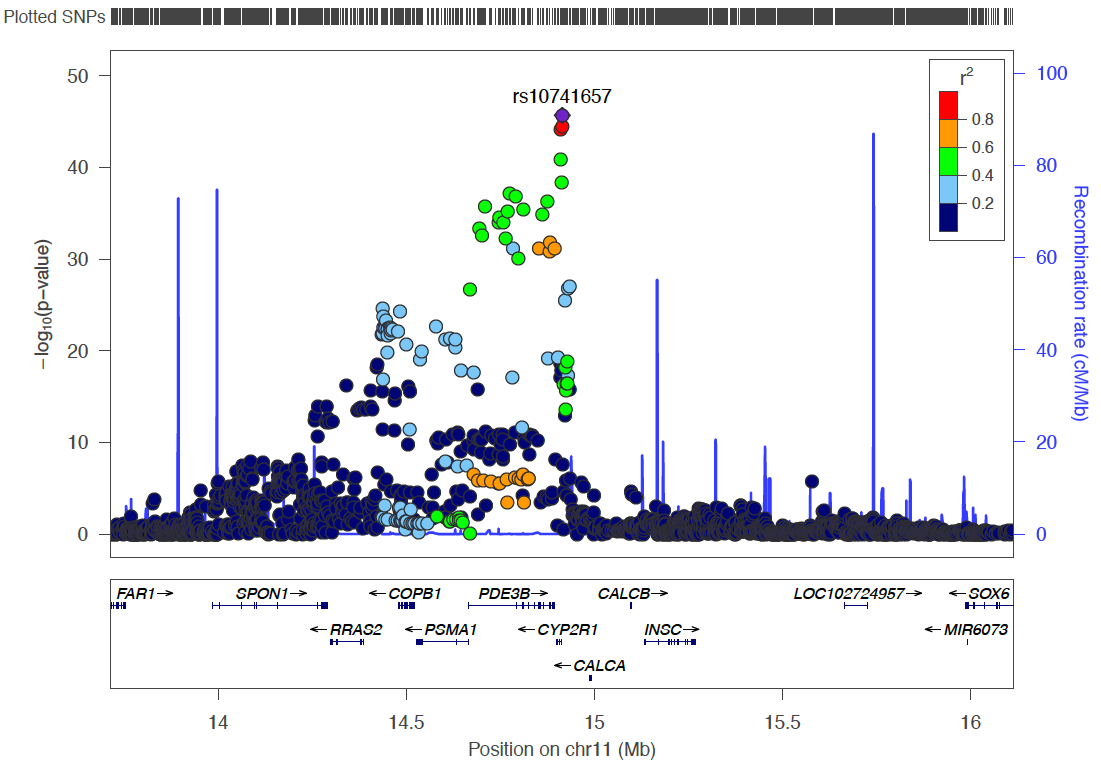
tmp <- data %>%
filter(ProbeChr==11) %>%
filter(Probe_bp>14000000 & Probe_bp<15500000) %>%
dplyr::select(Gene, topSNP, p_SMR, Probe_bp, topSNP_bp) %>%
gather(key, position, -1, -2, -3) %>%
mutate(position=position/1000000)
tmp %>% ggplot( aes(y=position, x=key)) +
geom_line(aes(group=paste(Gene,topSNP), color=ifelse(p_SMR<thres,"signif","non signif"))) +
geom_point() +
geom_text(
data=subset(tmp, key=="Probe_bp") %>% dplyr::select(Gene, key, position) %>% group_by(Gene) %>% summarize(key=unique(key), position=mean(position)),
aes( label=Gene) , angle=45, color="grey", hjust=1, nudge_x = -0.1
) +
geom_text(
data=subset(tmp, key=="topSNP_bp") %>% dplyr::select(topSNP, key, position) %>% group_by(topSNP) %>% summarize(key=unique(key), position=mean(position)),
aes( x=key, label=topSNP) , angle=45, color="grey", hjust=0, nudge_x = 0.1
) +
coord_flip() +
theme_ipsum() +
xlab("") +
ylab("Position (Mb)") +
labs(color="SMR assoc with Vitamin-D")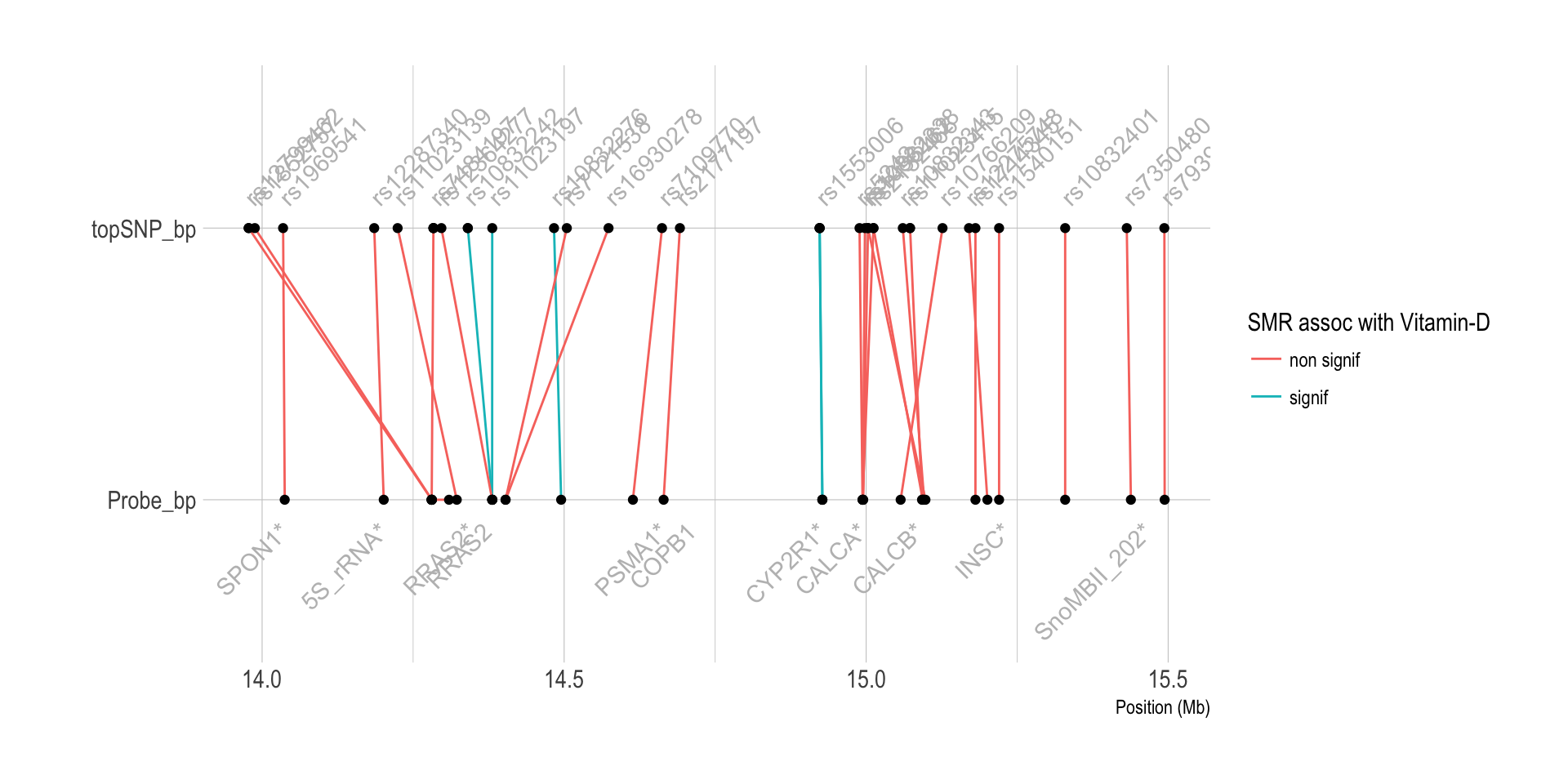
Chromo 11 ~ 71Mb (DHCR7)
# Show significant association:
data %>%
filter(p_SMR < thres & topSNP_chr==11 & topSNP_bp>40000000) %>%
arrange(Probe_bp)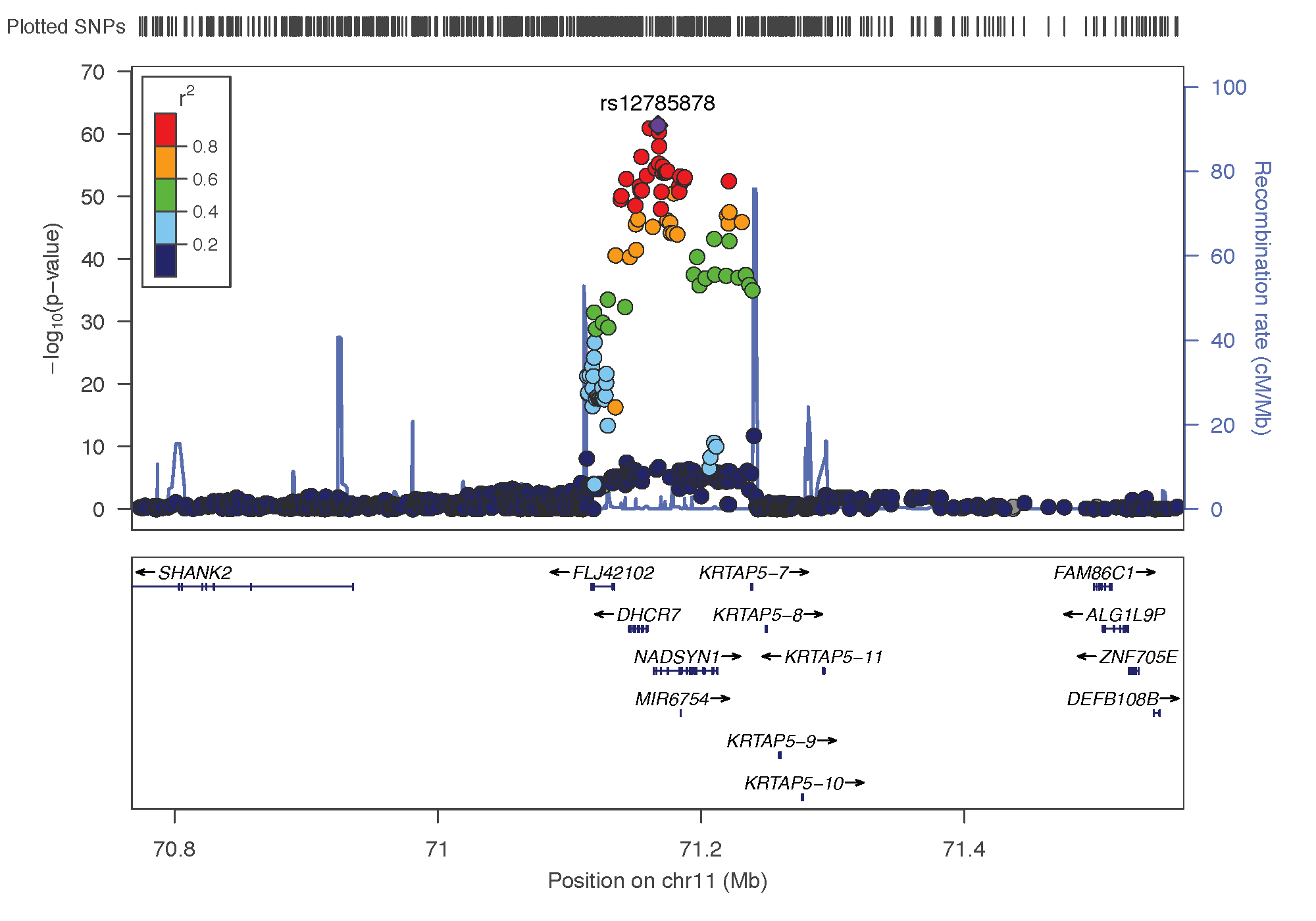
tmp <- data %>%
filter(ProbeChr==11) %>%
filter(Probe_bp>71000000 & Probe_bp<71500000) %>%
dplyr::select(Gene, topSNP, p_SMR, Probe_bp, topSNP_bp) %>%
gather(key, position, -1, -2, -3) %>%
mutate(position=position/1000000)
tmp %>% ggplot( aes(y=position, x=key)) +
geom_line(aes(group=paste(Gene,topSNP), color=ifelse(p_SMR<thres,"signif","non signif"))) +
geom_point() +
geom_text(
data=subset(tmp, key=="Probe_bp") %>% dplyr::select(Gene, key, position) %>% group_by(Gene) %>% summarize(key=unique(key), position=mean(position)),
aes( label=Gene) , angle=45, color="grey", hjust=1, nudge_x = -0.1
) +
geom_text_repel(
data=subset(tmp, key=="topSNP_bp") %>% dplyr::select(topSNP, key, position) %>% group_by(topSNP) %>% summarize(key=unique(key), position=mean(position)),
aes( x=key, label=topSNP) , angle=45, color="grey", hjust=0, nudge_x = 0.1
) +
coord_flip() +
theme_ipsum() +
xlab("") +
ylab("Position (Mb)") +
labs(color="SMR assoc with Vitamin-D")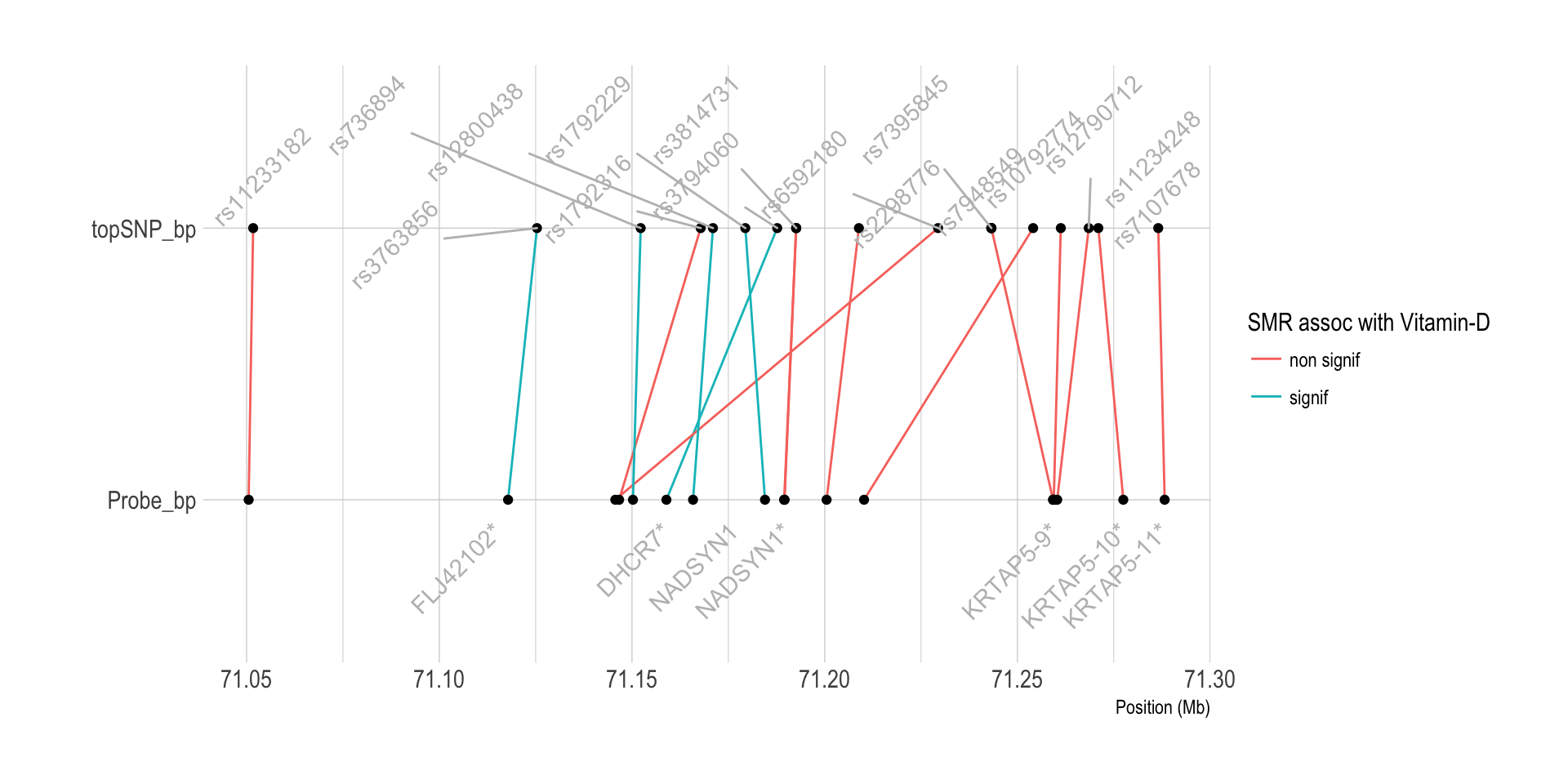
A work by Yan Holtz
Yan.holtz.data@gmail.com